Data Management
Tutorial
Users cannot modify the data in an OmixAtlas directly. Instead, all data management operations (i.e. creation, updation or deletion of datasets) are done through a Catalog. Each OmixAtlas has an internal data Catalog that acts as the source of truth for the data, and any modifications done to it are eventually propagated to the OmixAtlas.
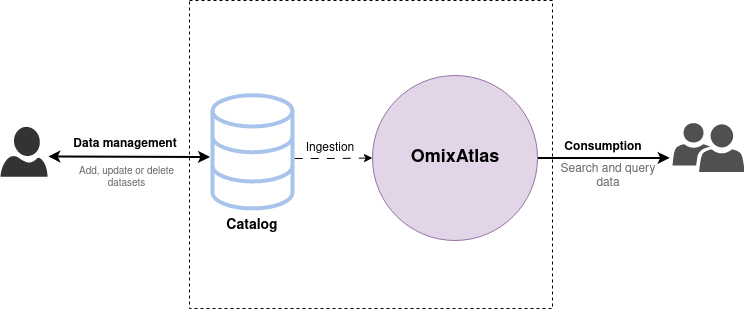
How do Catalogs work?
A Catalog is a collection of versioned datasets.
You can access it through the Catalog class.
from polly.auth import Polly
from polly.data_management import Catalog
Polly.auth("<access_key>")
catalog = Catalog(omixatlas_id="9")
Here's how you list the datasets inside the catalog.
catalog.list_datasets(limit=4)
# [
# Dataset(dataset_id='PXD056', data_type='Proteomics', ...),
# Dataset(dataset_id='SCP890', data_type='Single Cell RNASeq', ...),
# Dataset(dataset_id='GSE583_GPL8887', data_type='Bulk RNAseq', ...),
# Dataset(dataset_id='GSE8304', data_type='Bulk RNAseq', ...),
# ]
Each dataset is uniquely identified by a dataset_id and has some other attributes associated with it.
dataset = catalog.get_dataset(dataset_id='SCP890')
print(dataset)
# Dataset(dataset_id='SCP890', data_type='Single Cell RNASeq', ...),
print(dataset.data_type)
# 'Single Cell RNASeq'
Datasets consist of a metadata dictionary and a data file.
# load the metadata
dataset.metadata()
# {'description': 'Distinct transcriptional ...', 'abstract': 'PD-1 blockade unleashes CD8 T cells1, including...' }
# download the data into an h5ad file
dataset.download("./mydataset.h5ad")
# alternatively, load the data directly into memory
adata = dataset.load()
The data, metadata or data type of a dataset can be modified.
import scanpy
# load data
adata = catalog.get_dataset('SCP890').load()
# filter cells
scanpy.pp.filter_cells(adata)
# update data
catalog.update_dataset('SCP890', data=adata)
Creating a new dataset
You can create a new dataset in the catalog using the Catalog.create_dataset function.
new_dataset = catalog.create_dataset(
dataset_id='GSE123',
data_type='Bulk RNAseq',
data='path/to/data/file.gct',
metadata={'description': '...', 'tissue': ['lung']}
)
The metadata is passed as a dictionary. The data file (e.g. h5ad, gct) can be passed either by providing the path to the file or by passing the AnnData or GCToo object directly to the data parameter.
Passing ingestion parameters
The ingestion priority, ingestion_run_id and indexing flags are optional arguments that can be passed using the ingestion_params argument.
new_dataset = catalog.create_dataset(
dataset_id='GSE123',
data_type='Bulk RNAseq',
data='path/to/data/file.gct',
metadata=dict(),
ingestion_params={'priority': 'high', 'file_metadata': True, 'ingestion_run_id': '<run_id>'}
)
Versioning
Every call to catalog.update_dataset(...) creates a new version of the dataset.
You can list all prior versions of a dataset and access their underlying data.
versions = catalog.list_versions(dataset_id='GSE101720_GPL18573_raw')
for version in versions:
print(version.metadata())
Its also possible to roll back to an older version of a dataset.
Supplementary files
Supplementary files are arbitrary files that can be attached to a dataset. They can contain anything from the results of an analysis to raw sequence data.
dataset_id = 'GSE101720_GPL18573_raw'
catalog.attach_file(dataset_id, path='./analysis/report.html')
# SupplementaryFile(file_name='report.pdf', file_format='pdf', created_at='2024-01-17 14:39:48')
file_name. Uploading multiple files with the same name will cause previous ones to be overwritten
These files can be also be tagged with a string value. These "tags" can be used as a means to organize files into categories.
catalog.attach_file(dataset_id, './de_results.csv', tag='Diff Exp Results')
catalog.attach_file(dataset_id, './counts/vst_normalized_counts.csv', tag='Diff Exp Results')
Here's how you would download all the differential expression results uploaded above.
sup_files = catalog.list_files(dataset_id)
sup_files
# [
# SupplementaryFile(file_name='report.pdf', file_format='pdf', created_at='2024-01-17 14:39:48')
# SupplementaryFile(file_name='de_results.csv', file_format='csv', created_at='2024-01-17 14:39:48', tag='Diff Exp Results')
# SupplementaryFile(file_name='vst_normalized_counts.csv', file_format='csv', created_at='2024-01-17 14:39:48', tag='Diff Exp Results')
# ]
for file in sup_files:
if file.tag == "Diff Exp Results":
file.download('./')
Supplementary files are not versioned unlike datasets and cannot be retrieved once deleted.
API Reference
Catalog
create_dataset
Creates a new dataset in the catalog.
Raises an error if a dataset with the same ID already exists
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
dataset_id
|
str
|
The unique identifier for the dataset. |
required |
data_type
|
str
|
The type of data being uploaded (e.g. |
required |
data
|
Union[str, GCToo, AnnData]
|
The data, either as a file or as an AnnData or GCToo object |
required |
metadata
|
dict
|
The metadata dictionary |
required |
Returns:
| Type | Description |
|---|---|
Dataset
|
Newly created dataset. |
Examples:
get_dataset
Retrieves the dataset with the given dataset_id if it exists
This function doesn't download the underlying data, it only returns a reference to it
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
dataset_id
|
str
|
The unique identifier for the dataset to retrieve. |
required |
Returns:
| Type | Description |
|---|---|
Dataset
|
An object representing the retrieved dataset. |
Examples:
update_dataset
Updates one or more attributes of a dataset.
Every update creates a new version of the dataset.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
dataset_id
|
str
|
Identifier for the dataset. |
required |
data_type
|
Optional[str]
|
The type of data being uploaded (e.g. |
None
|
data
|
Union[str, GCToo, AnnData, None]
|
The data, either as a file or as an AnnData or GCToo object |
None
|
metadata
|
Optional[dict]
|
The metadata dictionary |
None
|
Returns:
| Type | Description |
|---|---|
Dataset
|
The updated dataset |
Examples:
delete_dataset
list_datasets
Retrieves the complete list of datasets in the catalog.
If prefetch_metadata is True the metadata json is downloaded in advance
using multiple threads. This makes the dataset.metadata() function call
return instantly. This is useful for bulk metadata download.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
limit
|
Optional[int]
|
Limits the number of datasets that are returned |
None
|
prefetch_metadata
|
bool
|
Prefetches metadata for each dataset |
False
|
Returns:
| Type | Description |
|---|---|
List[Dataset]
|
A list of Dataset instances. |
Examples:
__contains__
Check if a dataset with the given dataset_id exists in the catalog.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
dataset_id
|
str
|
Identifier for the dataset. |
required |
Returns:
| Type | Description |
|---|---|
bool
|
True if the dataset exists, False otherwise. |
set_parent_study
Adds a dataset to a study. The study_id can be any arbitrary string.
To remove the dataset from the study, set the study_id to None.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
dataset_id
|
str
|
Identifier for the dataset |
required |
study_id
|
Optional[str]
|
User defined ID for the parent study |
required |
Examples:
Add dataset to a study
Remove the dataset from the study
list_studies
list_datasets_in_study
Retrieves a list of datasets associated with a specific study.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
study_id
|
str
|
Identifier for the study. |
required |
Returns:
| Type | Description |
|---|---|
List[str]
|
A list of dataset IDs that are part of the study. |
Examples:
get_version
Retrieves a specific version of a dataset.
You can also use this to retrieve a version that may have been deleted.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
dataset_id
|
str
|
Identifier for the dataset. |
required |
version_id
|
str
|
Identifier for the dataset version. |
required |
Returns:
| Type | Description |
|---|---|
DatasetVersion
|
An object representing the dataset version. |
Examples:
list_versions
Lists dataset versions, optionally including deleted ones.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
dataset_id
|
str
|
Identifier for the dataset. |
required |
include_deleted
|
bool
|
If set to True, includes previously deleted versions |
False
|
Returns:
| Type | Description |
|---|---|
List[DatasetVersion]
|
List of dataset versions. |
rollback
Reverts a dataset to a specified older version.
This function updates the dataset's data and metadata to match the specified
version. The version ID itself is not restored.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
dataset_id
|
str
|
Identifier for the dataset. |
required |
version_id
|
str
|
Identifier for the version to roll back to. |
required |
Returns:
| Type | Description |
|---|---|
Dataset
|
The dataset instance with the rolled back data and metadata. |
attach_file
Attach a supplementary file to a dataset.
If a file with the same name already exists it is overwritten.
Optionally, you can link the supplementary file to a specific version of a dataset.
This will auto-delete the file if the underlying data for the dataset changes.
Note that changes to metadata or data_type will not auto-delete the file.
Added in polly-python v1.3
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
dataset_id
|
str
|
Identifier for the dataset. |
required |
path
|
str
|
Local path of the file. |
required |
tag
|
Optional[str]
|
An optional tag for the supplementary file. |
None
|
source_version_id
|
Optional[str]
|
The version_id of the source dataset. |
None
|
file_name
|
Optional[str]
|
The name of the file. If not provided the file name will be inferred from the path. |
None
|
Returns:
| Type | Description |
|---|---|
SupplementaryFile
|
A SupplementaryFile instance representing the attached file. |
get_file
Retrieves a supplementary file.
Added in polly-python v1.3
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
dataset_id
|
str
|
Identifier for the dataset. |
required |
file_name
|
str
|
Name of the file to retrieve. |
required |
Returns:
| Type | Description |
|---|---|
SupplementaryFile
|
A SupplementaryFile instance. |
list_files
Retrieves a list of all supplementary files attached to a dataset.
Added in polly-python v1.3
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
dataset_id
|
str
|
Identifier for the dataset. |
required |
Returns:
| Type | Description |
|---|---|
List[SupplementaryFile]
|
A list of SupplementaryFile instances. |
delete_file
Deletes the supplementary file.
Added in polly-python v1.3
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
dataset_id
|
str
|
Identifier for the dataset. |
required |
file_name
|
str
|
File name |
required |
clear_token_cache
Clears cached S3 tokens
trigger_ingestion
A helper function to manually trigger ingestion for a dataset
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
dataset_id
|
str
|
Identifier for the dataset. |
required |
Dataset
Bases: VersionBase
Class that represents a dataset
Attributes:
| Name | Type | Description |
|---|---|---|
dataset_id |
str
|
Dataset Identifier |
omixatlas_id |
str
|
OmixAtlas ID (e.g. |
data_type |
str
|
Data type (e.g. |
data_format |
str
|
Storage format for the data (e.g. |
version_id |
str
|
Unique version identifier |
study_id |
Optional[str]
|
Identifier for the parent study |
last_modified_at |
int
|
Unix timestamp (ms) for when the dataset was last modified |
download
load
DatasetVersion
Bases: VersionBase
Class that represents an immutable version of a dataset
Attributes:
| Name | Type | Description |
|---|---|---|
dataset_id |
str
|
Dataset Identifier |
omixatlas_id |
str
|
OmixAtlas ID (e.g. |
data_type |
str
|
Data type (e.g. |
data_format |
str
|
Storage format for the data (e.g. |
version_id |
str
|
Unique version identifier |
created_at |
int
|
Unix timestamp (ms) for when this version was created |
download
load
SupplementaryFile
Class that represents a supplementary file
Attributes:
| Name | Type | Description |
|---|---|---|
dataset_id |
str
|
Dataset Identifier |
omixatlas_id |
str
|
OmixAtlas ID (e.g. |
file_name |
str
|
Unique identifier for the file |
file_format |
str
|
Format of the file (e.g. |
tag |
Optional[str]
|
An optional tag for the file |
created_at |
int
|
Unix timestamp (ms) for when the file was added (or modified) |
copy_dataset
Copies a dataset from one omixatlas catalog to another.
Any supplementary files attached to the dataset are also copied.
The transfer happens remotely, the data is not downloaded locally.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
src_omixatlas_id
|
str
|
The source omixatlas_id |
required |
src_dataset_id
|
str
|
The source dataset_id |
required |
dest_omixatlas_id
|
str
|
The destination omixatlas_id |
required |
dest_dataset_id
|
str
|
The destination dataset_id |
required |
overwrite
|
bool
|
If True, overwrites the destination dataset if it already exists |
False
|
Returns:
| Type | Description |
|---|---|
Dataset
|
The dataset in the destination catalog. |
Examples:
(Advanced) Bulk operations
The Catalog class currently doesn't provide any built-in methods to do operations on a large number of datasets. However, you could make use of the code snippet shown below. It shows how to add a new field to the metadata of all datasets in a catalog.
# Download all metadata
datasets = catalog.list_datasets(prefetch_metadata=True)
# Create a mapping from dataset_id to the new metadata dictionaries
dataset_id_to_metadata = {}
for dataset in datasets:
new_metadata = {**dataset.metadata(), "new_field": "foobar"}
dataset_id_to_metadata[dataset.dataset_id] = new_metadata
# Import helper function
from polly.threading_utils import for_each_threaded
# Run this function across multiple threads
def fn(dataset):
dataset_id = dataset.dataset_id
catalog.update_dataset(
dataset_id, metadata=dataset_id_to_metadata[dataset_id]
)
for_each_threaded(items=datasets, fn=fn, max_workers=10, verbose=True)