Pre-configured Environment
Polly supports various notebook environments in the form of dockers to cater to the needs of different users. Each of the dockers is built according to various data analytic needs ranging from basic scripting, processing large data or training and testing of ML models. The menu to select the notebook environments will pop-up whenever you create or upload the notebook and opens it for the first time.
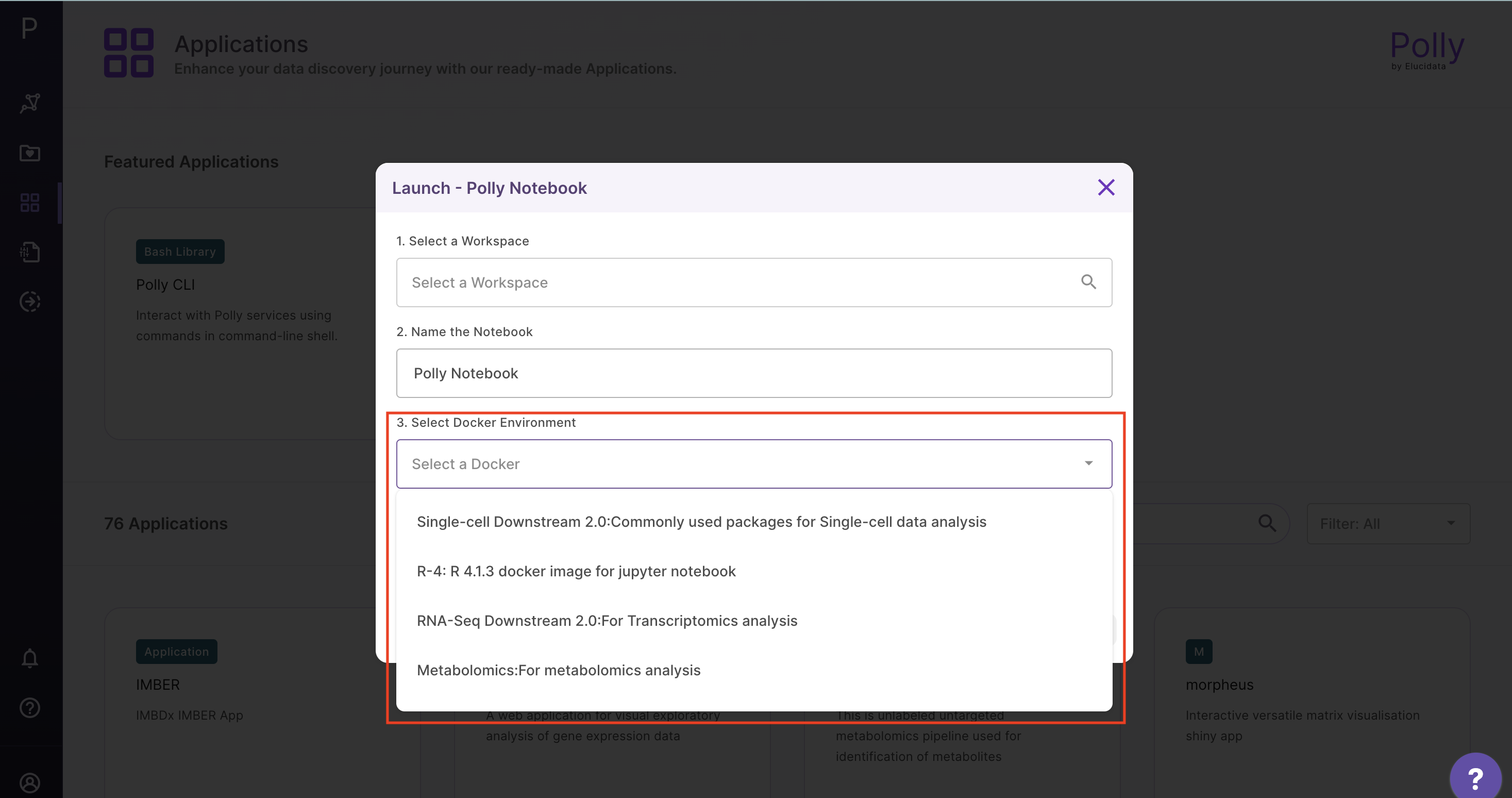
The various notebook environments supported are as follows:
| Environment | Usage | R libraries | Python Modules | System |
|---|---|---|---|---|
| R | General R scripting |
|
None | |
| Python 2 | General Python 2 scripting | None |
|
|
| Python 3 | General Python 3 scripting | None |
|
|
| Pollyglot | Multiple kernels (R, python and bash) in same notebook/environment |
|
|
|
| Barcoded Bulk RNA-seq | Alignment and processing of RNA-seq fastq files with barcodes |
|
|
|
| Machine Learning in python | Training, testing and validation of ML models | None |
|
|
| Single Cell Downstream | Single Cell Analysis |
|
|
|
| Data Exploration | R and python for general data analysis | All libraries from base R docker | All libraries from base python docker | |
| RNA-seq Downstream | Transcriptomics Analysis |
|
All libraries from base python docker | |
| Metabolomics | Metabolomics Analysis |
|
All libraries from base python docker |